paired end sequencing reads
A typical Illumina MiSeq sequencing run generates 20-30 million 2 300 bp paired-end sequence reads which roughly corresponds to 15 GB of sequence data to be processed. What is a paired-end read.

Paired End Sequencing Paired End Vs Single End Sequencing Pair End Reads Single End Reads Youtube
Paired-end vs single-end sequencing reads.
. Read files from paired-end sequencing need to be paired in Geneious before the pairing information can be used in assembly. Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data. Paired-end DNA sequencing reads provide high-quality alignment across DNA regions containing repetitive sequences and produce long contigs for de novo sequencing by filling gaps in the.
In general paired-end reads tend to be in a FR orientation have relatively small inserts 300 - 500 bp and are particularly useful for the sequencing of fragments that. When I read papers I find paired -end read and single-end reads are mentioned many times. But what is a.
Visit Maverix Biomics to learn more about RNA-seq. Paired-end sequencing facilitates detection of genomic. The term paired ends.
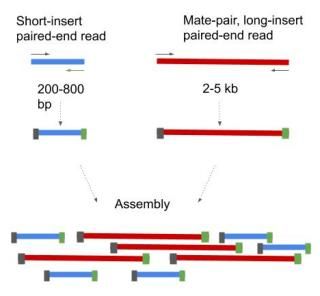
Based on the sequence of mimic integration of Gt1 gene a set of paired-end reads was generated randomly generated in silico using art-illumina software corresponding to the. Combining data from mate pair sequencing with those from short-insert paired-end reads provides increased information for maximising sequencing coverage across a genome 1. In paired-end reading it starts at one read finishes this direction at the specified read length and then starts another round of reading from the opposite end of the fragment.
The Illumina paired-end sequencing technology can generate reads from both ends of target DNA fragments which can subsequently be merged to increase. Paired-end DNA sequencing reads provide superior alignment across DNA regions containing repetitive sequences and produce longer contigs for de novo sequencing by filling gaps in the. RNA-seq analysis configuration on the Maverix Analytic Platform.
Paired-end reading improves the ability to identify the relative positions of various reads in the genome making it much more effective than single. In conventional paired-end sequencing you simply sequence using the adapter for one end and then once youre done you start over sequencing using the adapter for the other. The standard Illumina paired-end protocol produces reads oriented pointing toward each other just like good old fashioned Sanger paired reads but the insert size is much.
Another supposed advantage is that it leads to more accurate reads. Why is paired-end sequencing better. Paired-end DNA sequencing reads provide high-quality alignment across DNA regions containing repetitive sequences and produce long contigs for de novo sequencing by filling gaps in the.
This can be done using the Set Paired Reads. The 2 complementary DNA strands are oriented in opposite orientation and sequence reads from either end are generating results of those 2 different strands. One of the advantages of paired end sequencing over single end is that it doubles the amount of data.

Mapping And Further Processing Gbprocess 4 0 0 Documentation

What Is Mate Pair Sequencing For
![]()
Overview Of Amplicon Preprocessing
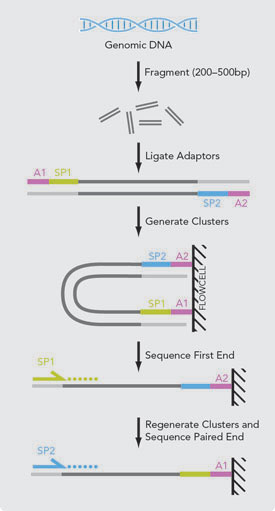
Paired End Sequencing Vs Mate Pair Sequencing Zhongxu Blog
Personal Genome Sequencing Current Approaches And Challenges
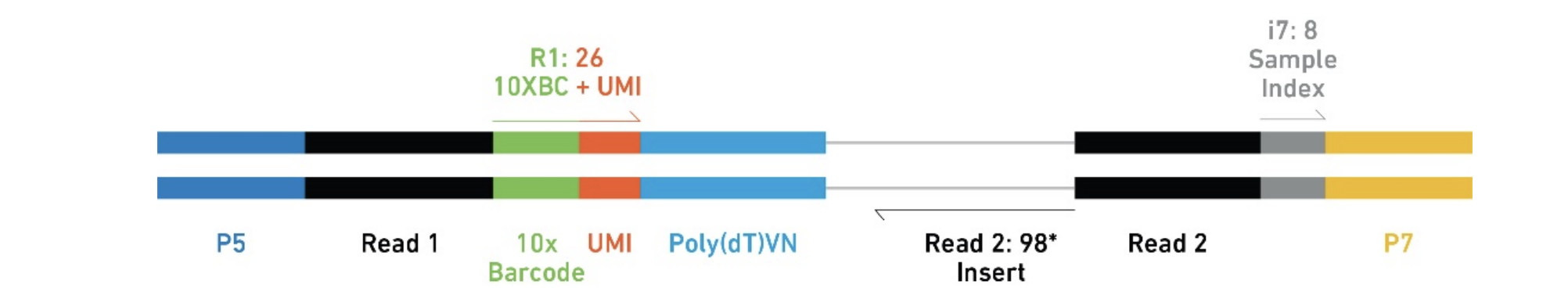
Understand 10x Scrnaseq And Scatac Fastqs Dna Confesses Data Speak
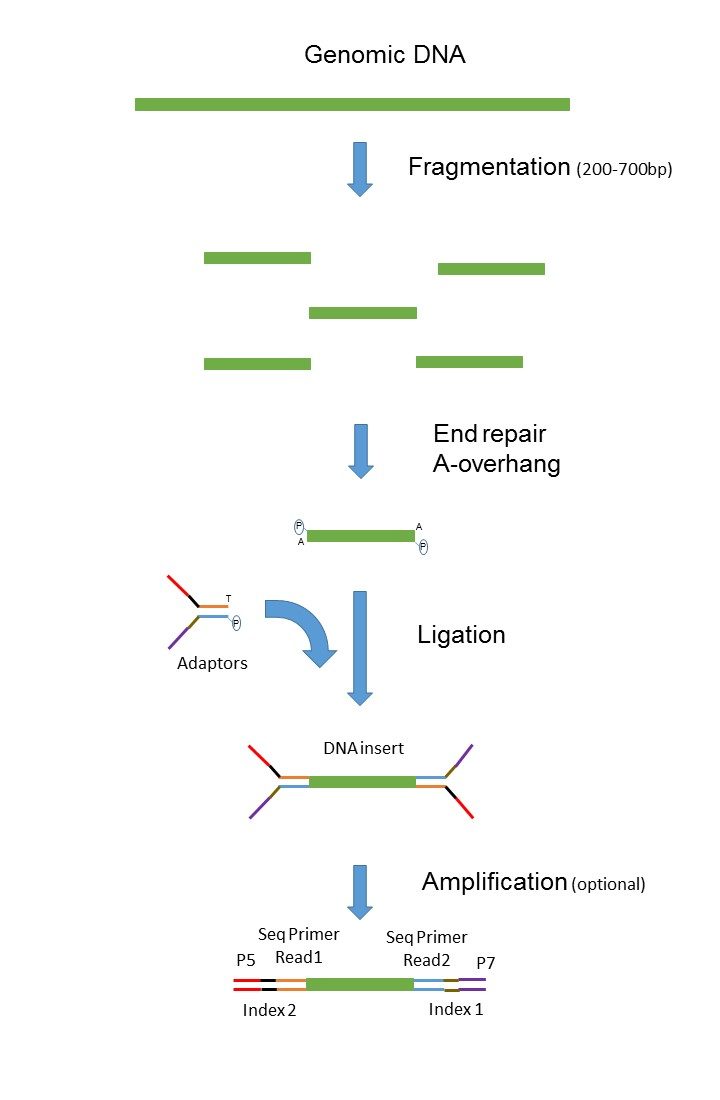
Short Read Sequencing Through Clustering France Genomique

What Is Mate Pair Sequencing For
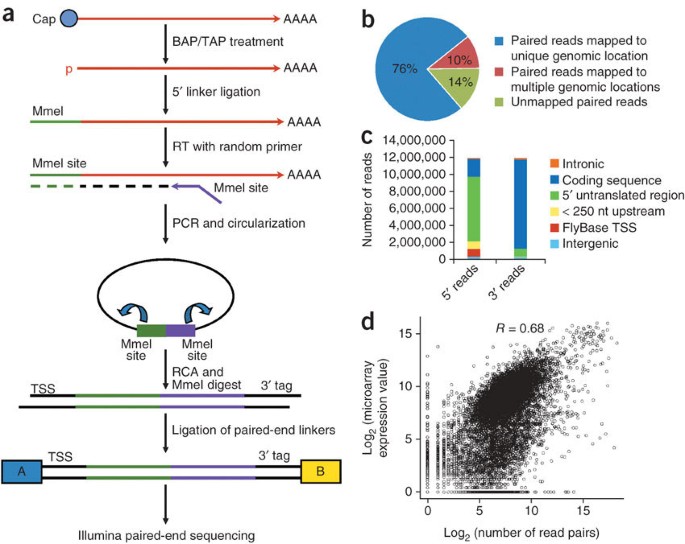
A Paired End Sequencing Strategy To Map The Complex Landscape Of Transcription Initiation Nature Methods
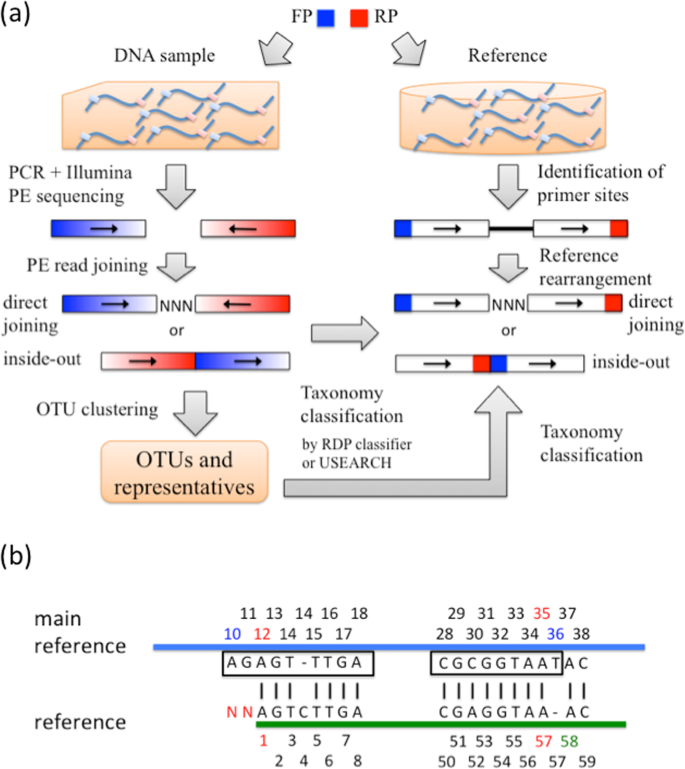
Joining Illumina Paired End Reads For Classifying Phylogenetic Marker Sequences Bmc Bioinformatics Full Text

Paired Vs Unpaired Dna Sequencing
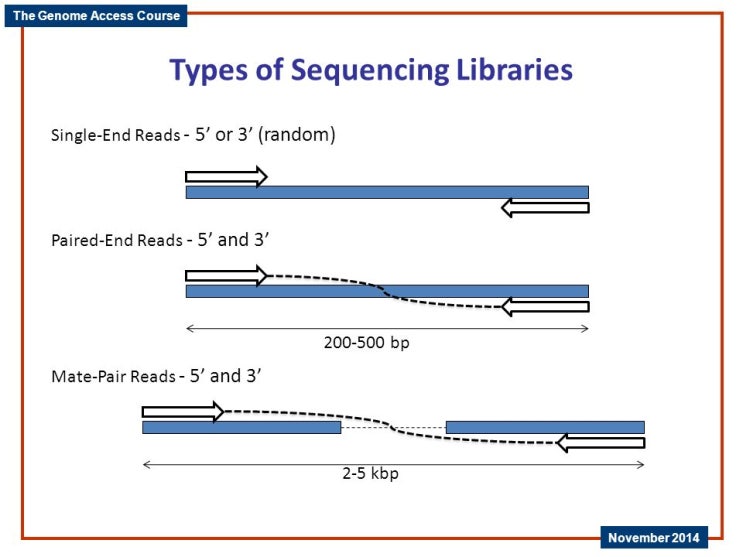
Single End Paired End Mate Pair 네이버 블로그
Pre Processing Paired End Illumina Data For Qiime Bioinformatics In Biomed
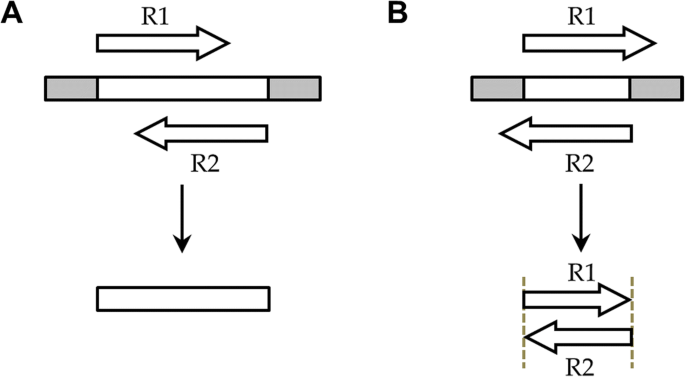
Ngmerge Merging Paired End Reads Via Novel Empirically Derived Models Of Sequencing Errors Bmc Bioinformatics Full Text
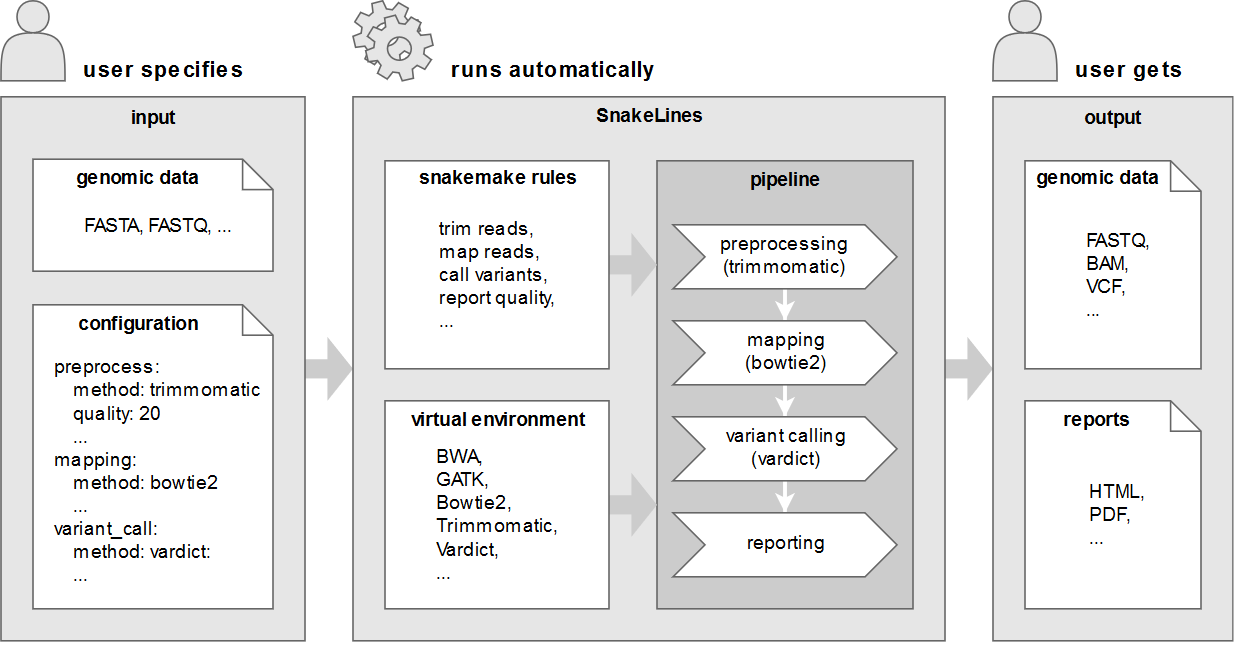
Snakelines Integrated Set Of Computational Pipelines For Paired End Sequencing Reads Geneton
Introduction To Genome Assembly Tutorial

Long Fragments Achieve Lower Base Quality In Illumina Paired End Sequencing Biorxiv
